pepFunk, an R shiny app
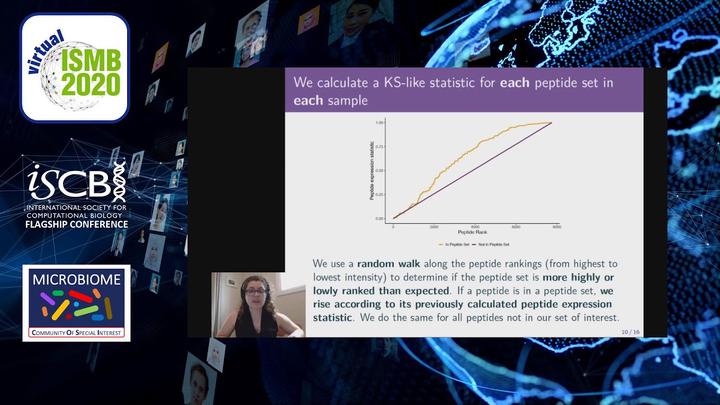 Image credit: YouTube
Image credit: YouTubeAbstract
Researchers can use metaproteomics to study the composition and functional contributions of the gut microbiome to human health. These metaproteomic data are acquired by a multistep process, first starting with enzymatic digestion of microbial proteins into smaller and more easily detectable peptides. These peptides are then processed by a mass spectrometer, and the obtained mass spectra are matched back to a peptide database. Typically, the metaproteomic data analysis pipeline involves the identification of each peptide to a potential parent protein. Challenges to unambiguous protein identification arise due to the nature of enzymatic digestion, where peptides can match back to multiple parent proteins. We developed pepFunk, a peptide-centric functional analysis of metaproteomic data methodology and tool to circumvent this challenge. We created a gut microbiome peptide-to-KEGG database and developed a functional enrichment strategy for peptide-level data. Our peptide-centric approach gives an enhanced ability for users to observe the biological processes taking place in the microbiome. Our tool is open source and is available as a Shiny web application at https://shiny.imetalab.ca/pepFunk.